In this lab exercise you will be introduced to the range of animal body plans and relationship of morphology to animal phylogenies and classifications. Historically, biologists relied upon morphological features, developmental patterns, and fossils to guide their thinking about evolutionary relationships. Since the advent of modern molecular biology techniques, biologists have been able to test many of the hypotheses based on morphology against the data gleaned from gene sequences and gene products (e.g., RNA & proteins). Some of our morphologically based hypotheses have been supported by this evidence, while others have not (recall our last lecture). Today we will explore the kinds of morphological evidence observed from animal body plans and how this data can be applied to help deduce evolutionary relationships. We will then use these morphological characteristics to classify animals of unknown phylogenetic affiliation using a dichotomous key to the animal phyla.

Biologists organize life’s diversity within a system of hierarchical groupings, or taxa, that reflect their evolutionary relationships (i.e., phylogeny: phylum is the level in the taxonomic scheme that corresponds to what we are calling a body plan). Systematics is the study of biological diversity in an evolutionary context. It includes taxonomy, which is the naming and classification of species and groups of species. The 18th century scientist Linnaeus developed the hierarchical classification system that is the basis of our current naming and classification system (i.e., the species binomial and the domain, kingdom, phylum, class, order, family, genus, species hierarchy). Although he predated Darwin, his system of organization is well suited to representing evolutionary relationships because of its hierarchical structure. However, many biologists today favor modifying this scheme to one with a less rigid structure of ranks that may more naturally reflect evolutionary relationships and branching patterns (e.g., PhyloCode). This is an ongoing debate in the field and is not currently resolved.
People interested in identifying species usually refer to a key that is based on morphological characteristics of the organism. Scientists typically use a technical version of a key called a taxonomic (or dichotomous) key to identify species. Taxonomic keys can be simple, like those found in field guidebooks used by naturalists or interested readers or they may be highly technical documents used by scientists to identify unknown species. Because taxonomic keys typically use only morphological features, they may or may not reflect evolutionary history.
Objectives:
- Learn concepts and vocabulary associated with describing animal body plans.
- Learn about unique larval forms and features associated with different phyla
- Apply what you’ve learned and use a dichotomous key to classify unfamiliar organism into their respective phyla.
Background
Modern taxonomy uses cladistic methods as a primary way to group organisms into phylogenetic trees; the main goal is to accurately reflect evolutionary relationships among organisms, rather than morphological diversity. Cladistic methods for generating phylogenetic trees can incorporate many kinds of data, including fossil evidence, morphological features, developmental characteristics, and molecular data (gene sequences). Because Linnean taxonomic classifications were developed before the advent of molecular methods, many long-established taxonomic groupings may not accurately reflect our current understanding of evolutionary relationships. Any phylogenetic tree or classification must be viewed as a hypothesis, and therefore is subject to re-evaluation whenever new evidence comes to light.
While this is good science, it can be very frustrating for students (and scientists alike!), who must strive to understand both older and more recent classifications.
There are two primary questions one must try to answer in developing a classification system that reflects evolutionary relationships, regardless of the kinds of evidence used: 1) are shared traits among organisms the result of a common evolutionary origin, or convergent evolution (are the characters homologous or analogous)? and 2) what was the direction of evolutionary change (polarity)?
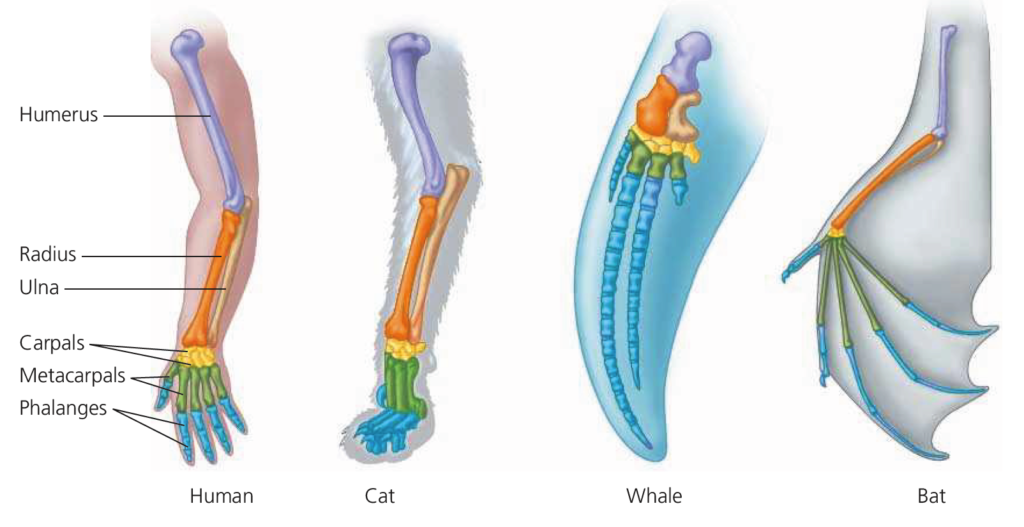
adapted for different functions, the forelimbs of all mammals are constructed from the same basic skeletal elements: one large bone (purple), attached to two smaller bones (orange and tan), attached to several small bones (gold), attached to several metacarpals (green), attached to approximately five digits, each of which is composed of multiple phalanges (blue).
Similar appearances are sometimes, but not always, reliable indicators of evolutionary relationships. Consider Ornithorhynchus, the weird duck-billed platypus, a semiaquatic animal from Australian and Tasmania. It has hair and milk glands like a mammal, a rubbery bill and webbed feet like a water bird, and lays leathery, shelled eggs in burrows like many reptiles. Within which group should the platypus be grouped? Which traits are most ‘important’ in an evolutionary context? During the 1800s the platypus was alternately placed in different groups (or considered an elaborate prank) as systematists argued these points. Today it is agreed that the presence of hair and milk glands indicates that the platypus is a primitive type of mammal. The available evidence suggests that hair and milk glands were passed down from a common ancestor and that these traits are thus homologous with hair and milk glands in other mammals, while the webbed feet are the result of convergent evolution.
Other examples of analogous structures are the wings of pterosaurs (extinct flying reptiles), and those of birds and bats. Note that the forelimbs are considered homologous structures but the wing structures themselves are analogous. Clearly determining the differences can be a very challenging task! In many cases, but not all, molecular evidence has supported major taxonomic groupings and proposed lineages derived from the study of morphological features. In this exercise you will become familiar with the classical concepts used to compare anatomical traits of organisms and seek to identify evolutionary relationships.
How to Use a Taxonomic Key
Figure 1. Example of homology and convergence. Pterosaurs, birds, and bats independently evolved adaptations of the vertebrate forelimb for flight. The bones of the arm in each are homologous, while specialized structures (feathers or wing membranes) evolved separately (convergent evolution).

How to use a Taxonomic Key:
A taxonomic key is a tool that is used to identify different types of organisms. An example of a key that could be used to identify a set of microorganisms is provided below. A taxonomic key contains a series of statements that describe the traits of the organisms, as shown in the sample key presented below. The statements are grouped into alternative descriptions (such as 1a and 1b) for each trait. To use the key, traits of the unknown organism are identified while following the steps of the key. When the description in the key and trait of the organism match, you are instructed to proceed to another set of descriptions or given an identification.
Correctly following the steps of the key eventually leads to identification of the unknown organism. Your lab instructor will walk you through an example.